How to use the data collected as knowledge?
Like I have written before, we need to rethink the binary data representation that we have currently and evaluate the effectiveness of it for an AI application. My take is that we should not collect the raw data as a binary encoded discrete scalar value periodically, but we need to use the data directly from the source and collect knowledge encoded in different forms either as a molecular structural form as we find in protein creation. As I have written in the various blogs on data (data continuity, missing depth of data), collecting raw data as scalar loses information that is critical for a knowledge and intelligence based system. When we collect knowledge rather than data, using the accumulative nature of some of the concepts already present in nature around us, the various relations between different parameters, their continuity and relative changes get automatically encoded in the formation. Thus, we do not have to write complex algorithms to relate discrete scalar values after the fact.
When we do this, we need to start re-thinking how this knowledge can be used? As I have written before, we need to start re-thinking all our presumptions on what “input”, “output”, “logic” etc is. In the computer world, input is anything that comes in a binary format, can be from a keyboard, network, IoT sensor, mouse clicks on a UI screen etc. As I have said in “Logic in AI“, for us logic is some pseudo-code that executes a set of instructions on a processor. But, this need not be so. If I am able to string together a series of molecular reactions to achieve a certain output, it becomes logic. Truly, even if I did not string them together in a sequence, but created a map, where “a protein” is created, and as it passes from one cell to the next “the appropriate peptide bond is broken breaking the protein” and the two pieces are transmitted in different paths, while the cell itself reacts correctly to the protein, a logic is achieved. For example, when we look at the “signal transduction” process, we find it does just such a logic.
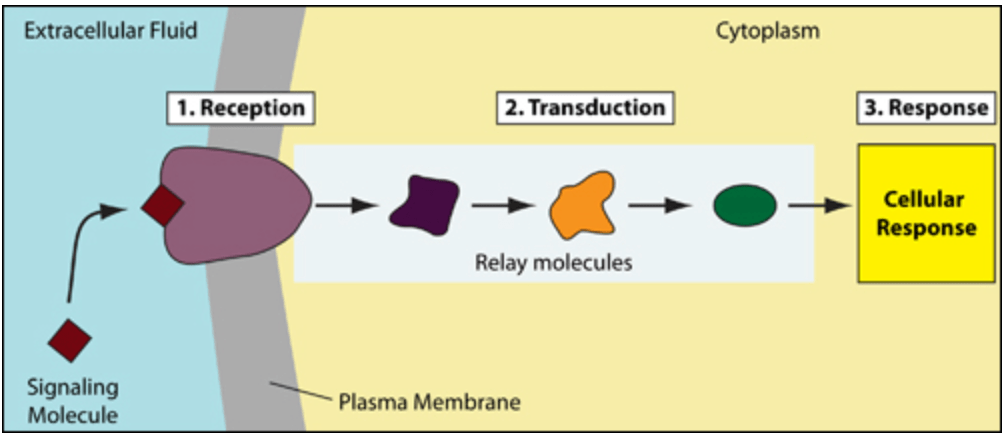
Signal transduction receives inputs using receptor proteins which is then transmitted via relay molecules to the cell that responds to the reactor protein. The simple image from libretexts:
If we read the various cellular responses that are present for a receptor, we find that some of the functions involve gene expression, which is turning on and off some specific gene. The internal protein is created using mRNA that are transcribed from these DNA. Thus, the internal proteins created are dependant on the data that is collected by the receptor proteins via logic that is encoded in the various molecules involved in the process of signal transduction. Here, we find that the logic has used “knowledge” without losing any of the continuity or depth of the data that is present for it to process. It should also be easily seen that the process while achieving a very complex logic, does not actually need huge processing power of a single processor or huge amounts of code to be written.
If we look at it from the perspective of the most common requirement that is currently present in AI, namely, image processing. In the current computer system, the image is looked upon as a 3D matrix of pixel values, the first and second are the image’s width and height, and the third dimension is the RGB values. Over this matrix, a sliding filter of some specific stride is used to compute the dot product of the filter matrix to the image matrix at the overlap window as the filter slides along with the image. The output then indicates a pattern, which is then used to identify the features in the image. I have not gone into all the implementation details, nor will I go into the problems in such a process. But, it should be mentioned that information can be lost based on the filter, the stride and the resolution of the image. The most crucial point to note here is that the image is static.
If, rather than doing this, i.e., convert the light stream to a static image at a given instant and then process the grid as pixels, say we left data for what it was initially, which is a light stream. Further, let us say we can take this light stream and focus it via a lens on photo-sensitive molecules that react to various frequencies and intensity of light in a specific manner that we can track and make use of, such as absorption of photons to create a molecular structure (similar to the photoreceptors in the eye where a molecular reaction is triggered by the light to alter amino acid structures). Here, the same light used to create the image gets encoded in the molecular properties of photoreceptors. This gives us an encoding of the image as a molecular structure, which includes the continuity of change of scenery for a duration and is not just a static image. This would form many such molecular structures simultaneously encoding the change in the scene for the given time over the visible range.
If this can be transmitted without losing the information via molecular structures, then the correct cleaving of the molecular structure, and the appropriate usage of the molecules should directly create intelligence from it, rather than spend crucial processing resources to understand the image from a 2D projection.
I definitely believe, the system we need to model for a knowledge and intelligence system needs to be similar to this, where data can be used directly from the source and converted via a series of reactions (i.e., direct usage for achieving an output) that do not lose information in the process of storing, retrieving and processing data.
Published on Java Code Geeks with permission by Raji Sankar, partner at our JCG program. See the original article here: How to use the data collected as knowledge? Opinions expressed by Java Code Geeks contributors are their own. |





Check https://www.wikipedia.org/